11 Bringing it All Together
library(mmir)
library(tidyverse)
library(rsample)
library(recipes)
library(parsnip)
library(workflows)
library(yardstick)
library(themis)Prepare the data for metric calculation.
11.1 Metric Calculation
The following code-chunk will produce more than 1,000 biological metrics.
metrics.df <- nest.df %>%
dplyr::mutate(
rich_family = taxa_rich(.dataframe = .,
.key_col = uid,
.group_col = family,
.unnest_col = data),
rich_genus = taxa_rich(.dataframe = .,
.key_col = uid,
.group_col = genus,
.unnest_col = data),
rich_target_taxon = taxa_rich(.dataframe = .,
.key_col = uid,
.group_col = target_taxon,
.unnest_col = data),
gini_simpson_ept = taxa_div(.dataframe = .,
.key_col = uid,
.counts_col = total,
.group_col = target_taxon,
.filter = order %in% c("ephemeroptera",
"plecoptera",
"trichoptera"),
.job = "gini_simpson",
.unnest_col = data),
simpson_ept = taxa_div(.dataframe = .,
.key_col = uid,
.counts_col = total,
.group_col = target_taxon,
.filter = order %in% c("ephemeroptera",
"plecoptera",
"trichoptera"),
.job = "simpson",
.unnest_col = data),
shannon_ept = taxa_div(.dataframe = .,
.key_col = uid,
.counts_col = total,
.group_col = target_taxon,
.filter = order %in% c("ephemeroptera",
"plecoptera",
"trichoptera"),
.job = "shannon",
.base_log = 2,
.unnest_col = data),
pct_ept = taxa_pct(.dataframe = .,
.key_col = uid,
.counts_col = total,
.filter = order %in% c("ephemeroptera",
"plecoptera",
"trichoptera"),
.unnest_col = data),
pct_cote = taxa_pct(.dataframe = .,
.key_col = uid,
.counts_col = total,
.filter = order %in% c("coleoptera",
"odonata",
"trichoptera",
"ephemeroptera"),
.unnest_col = data),
dom_1_target_taxon = taxa_dom(.dataframe = .,
.key_col = uid,
.counts_col = total,
.group_col = target_taxon,
.dom_level = 1,
.unnest_col = data),
dom_5_target_taxon = taxa_dom(.dataframe = .,
.key_col = uid,
.counts_col = total,
.group_col = target_taxon,
.dom_level = 5,
.unnest_col = data),
tol_index = taxa_tol_index(.dataframe = .,
.key_col = uid,
.counts_col = total,
.tol_col = ptv,
.unnest_col = data)
) %>%
dplyr::bind_cols(
taxa_seq(.dataframe = .,
.key_col = uid,
.counts_col = total,
.filter_cols_vec = c("class", "order", "family",
"ffg", "habit", "voltinism"),
.group_col = target_taxon,
.job = "rich",
.unnest_col = data),
taxa_seq(.dataframe = .,
.key_col = uid,
.counts_col = total,
.filter_cols_vec = c("class", "order", "family",
"ffg", "habit", "voltinism"),
.group_col = target_taxon,
.job = "pct_rich",
.unnest_col = data),
taxa_seq(.dataframe = .,
.key_col = uid,
.counts_col = total,
.filter_cols_vec = c("class", "order", "family",
"genus", "ffg", "habit", "voltinism"),
.job = "pct",
.unnest_col = data),
taxa_seq(.dataframe = .,
.key_col = uid,
.counts_col = total,
.filter_cols_vec = c("class", "order", "family",
"ffg", "habit", "voltinism"),
.group_col = target_taxon,
.job = "simpson",
.unnest_col = data),
)11.2 Metric Selection
11.2.1 Train/Test Split
set.seed(42)
train_test_split <- metrics.df %>%
select(-data) %>%
filter(!rt_nrsa_cat %in% "intermediate disturbance") %>%
mutate(rt_nrsa_cat = str_replace_all(rt_nrsa_cat, " ", "_"),
rt_nrsa_cat = factor(rt_nrsa_cat, levels = c("most_disturbed",
"least_disturbed"))) %>%
initial_split()
train.df <- training(train_test_split)
test.df <- testing(train_test_split)11.2.2 Preprocess
rec_obj <- recipe(rt_nrsa_cat ~ ., data = train.df) %>%
update_role(uid, new_role = "sample_id") %>%
step_normalize(all_predictors()) %>%
step_nzv(all_predictors()) %>%
step_corr(all_predictors(),
threshold = 0.75,
method = "spearman") %>%
step_pca(all_predictors(),
num_comp = 4) %>%
step_adasyn(rt_nrsa_cat,
neighbors = 5) %>%
prep()11.2.3 Model
## == Workflow [trained] ==========================================================
## Preprocessor: Recipe
## Model: logistic_reg()
##
## -- Preprocessor ----------------------------------------------------------------
## 5 Recipe Steps
##
## * step_normalize()
## * step_nzv()
## * step_corr()
## * step_pca()
## * step_adasyn()
##
## -- Model -----------------------------------------------------------------------
##
## Call: stats::glm(formula = formula, family = stats::binomial, data = data)
##
## Coefficients:
## (Intercept) PC1 PC2 PC3 PC4
## -0.94614 -0.54845 0.11392 0.02437 -0.01808
##
## Degrees of Freedom: 99 Total (i.e. Null); 95 Residual
## Null Deviance: 138.6
## Residual Deviance: 60 AIC: 7011.2.3.1 Predictions
pred_train <- predict(fit_train, new_data = train.df, type = "prob") %>%
bind_cols(select(train.df, rt_nrsa_cat))
pred_test <- predict(fit_train, new_data = test.df, type = "prob") %>%
bind_cols(select(test.df, rt_nrsa_cat))ggplot(pred_test, aes(.pred_least_disturbed, as.numeric(rt_nrsa_cat) - 1)) +
geom_point() +
stat_smooth(
method = "glm",
se = FALSE,
method.args = list(family = binomial))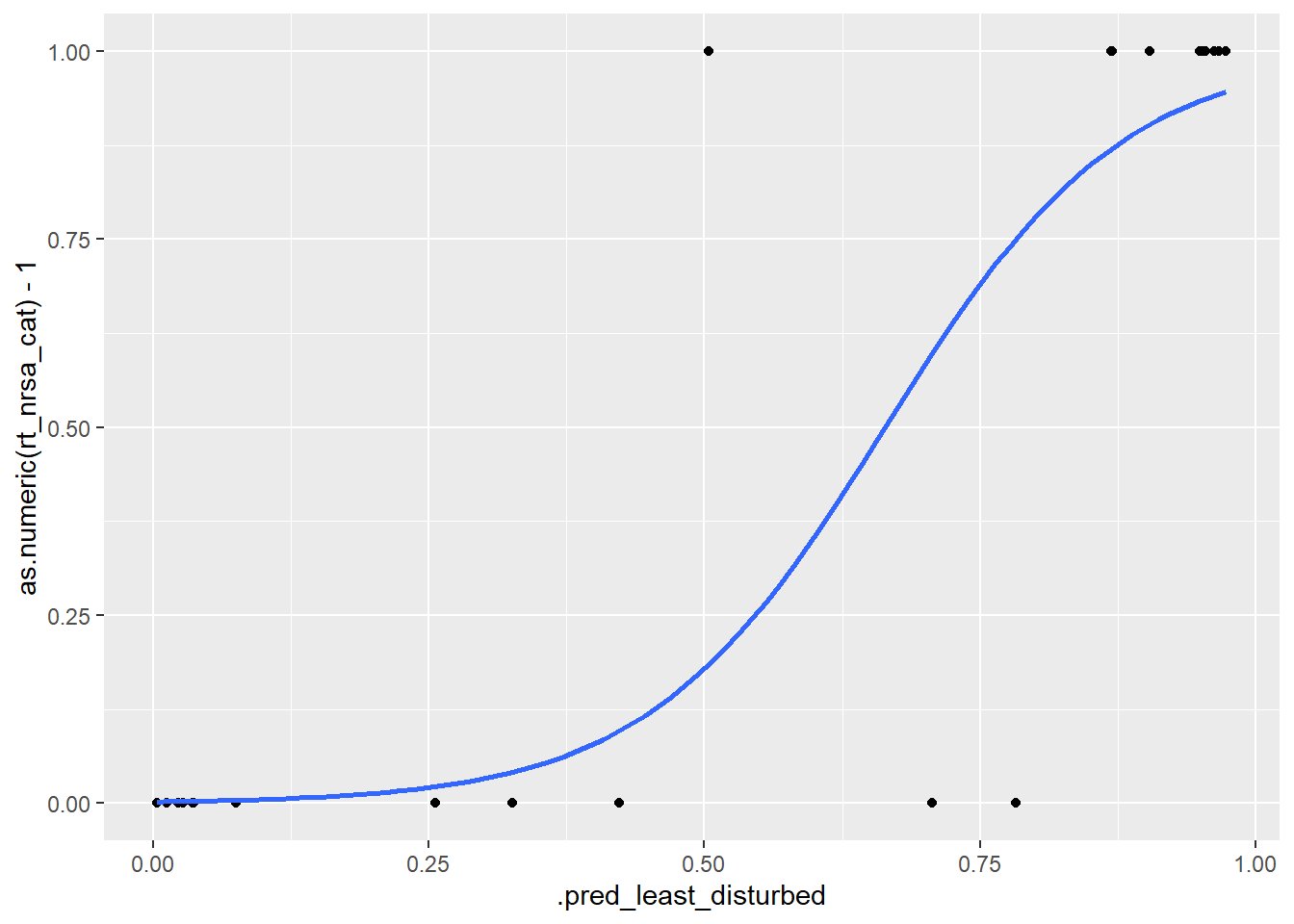
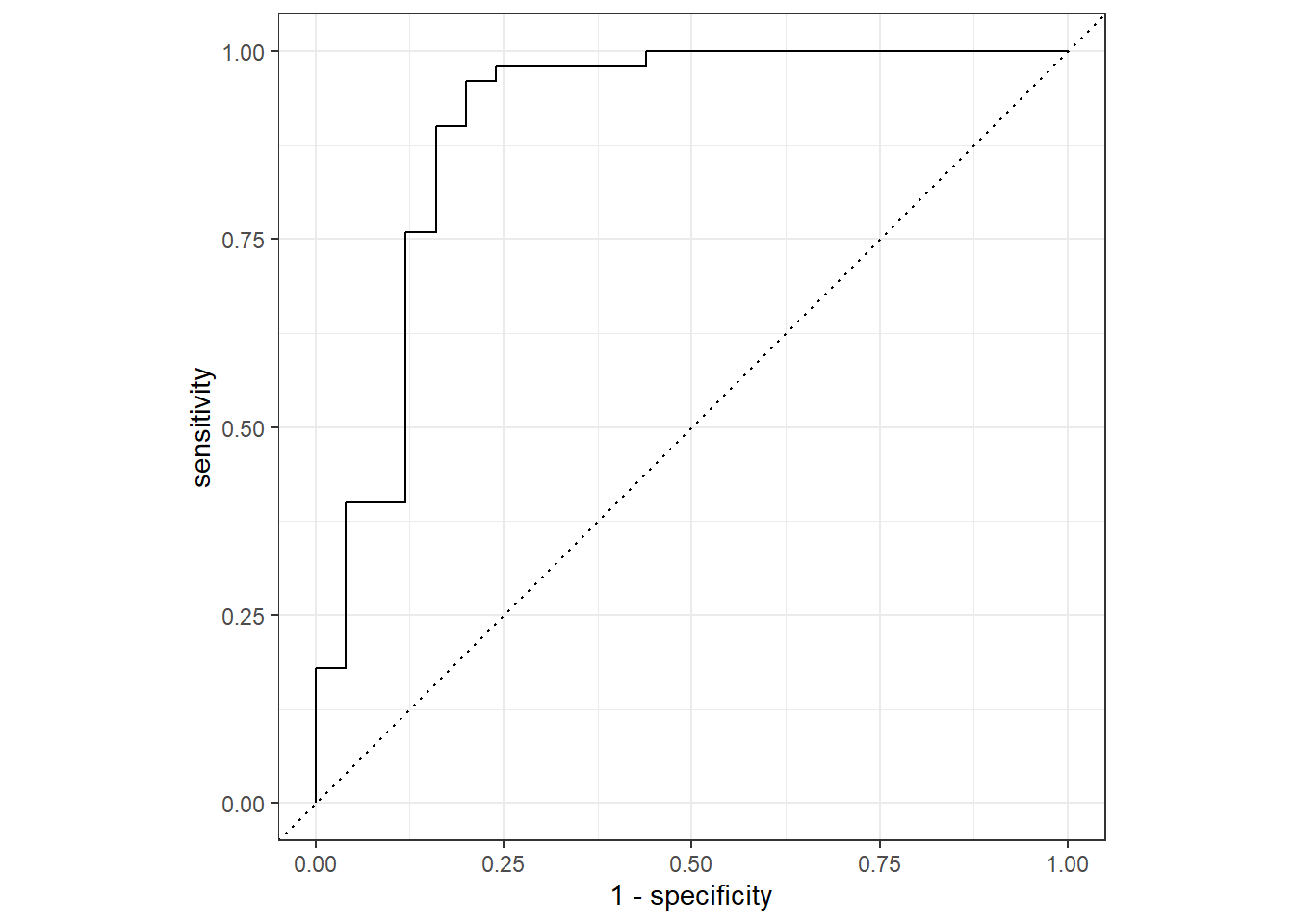
simple_glm_roc <- pred_train %>%
roc_curve(rt_nrsa_cat, .pred_least_disturbed)
pred_train %>%
roc_auc(rt_nrsa_cat, .pred_least_disturbed)## # A tibble: 1 x 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 roc_auc binary 0.9